DNA Barcoding: A Molecular Approach to Species Identification
Written by: Akhil Narayanan
DNA barcoding, first proposed by Paul Herbert in 2003, is a way to identify species by looking at certain sections of their DNA. Herbert’s work at the University of Guelph established the process by using a short DNA sequence, called the Cytochrome c oxidase I gene (COI). The COI gene varies a lot between species, making it possible to identify a variety of species through their DNA barcode. Let’s take a further look into DNA barcoding and its potential limitations.
What is DNA Barcoding?
DNA barcoding is a process that analyzes a section of the Cytochrome c oxidase I (COI) gene. The COI gene is used because it shows great variety between species known as interspecies variation, while also remaining stable within a species referred to as intraspecies variation. The difference between these two types of variation is known as a barcode gap, and the COI gene provides an optimal barcode gap for distinguishing animals with both accuracy and consistency. In a study where 64,000 insects’ COI gene was read, 78%–88% of the insects showed low intraspecies variation (Zhang et al., 2022). Similarly, an experiment using bacteria Wolbachia and other fungi symbionts revealed to researchers that using the COI gene could accurately identify 71 out of the 72 samples (Yang et al., 2020).Thus, the COI gene emerges as an ideal marker for which DNA can be identified.
The whole process of DNA barcoding starts from a sample of the specimen, such as hair, feathers or saliva. Methods such as organic extraction are used to extract the DNA out of the sample. Scientists use primers, nucleic acid sequences that act as a starting point for DNA synthesis, to target the COI gene. They then process the gene using PCR amplification, which creates multiple copies, making the gene more detectable and readable. Lastly, the COI gene undergoes DNA sequencing to determine the order of nucleotides that it consists of, effectively giving it its barcode with which it is identified (Kimball, 2016). Databases online, such as the Barcode of Life Data System (BOLD) and the NCBI nucleotide (nt) database, have a collection of COI sequences that can be compared with to identify millions of species.
Figure 1
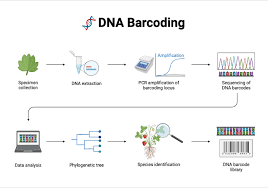
A Visual Showing the Steps of DNA Barcoding
Source: bioRender
What are some limitations to DNA Barcoding?
While much of the identification process is computerized, humans still play a part in DNA barcoding. They have to enter new specimens into the database. This creates a margin of error, since humans are prone to making mistakes like incorrectly labeling species before sequencing, confusing samples, and inappropriately using fixed distance thresholds (Chang et al., 2023). This is a considerable problem, because once a mislabeled sequence enters a database like the Barcode of Life Data System (BOLD), it causes problems for anyone else who tries to compare it with their own sample. These errors can lead to widespread misidentification, skew biodiversity assessments, and compromise conservation efforts that rely on accurate species data.
To address the limitations of DNA barcoding, especially the ones caused by human error, researchers have found novel technologies and better practices to improve DNA barcoding. For example, needing entries to be verified on databases. This reduces the errors humans commit when entering sequences. In addition, requiring standardized lab procedures reduces lab variability and error.
Figure 2
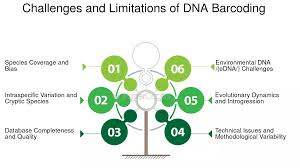
A Visual Showing the Different Challenges and Limitations of DNA Barcoding
Source: FasterCapital
Conclusion
DNA barcoding which was created by Paul Herbert. It uses the COI gene to accurately and reliably identify species, though human error and technical limitations remain. However, evolving technologies such as nanopore sequencing and better practices continue to improve and expand its potential.
References and Sources
19.1.9: Barcoding. (2016, August 3). Biology LibreTexts. https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Biology_%28Kimball%29/19%3A_The_Diversity_of_Life/19.01%3A_Eukaryotic_Life/19.1.09%3A_Barcoding
Antil, S., Abraham, J. S., Sripoorna, S., Maurya, S., Dagar, J., Makhija, S., Bhagat, P., Gupta, R., Sood, U., Lal, R., & Toteja, R. (2022). DNA barcoding, an effective tool for species identification: a review. Molecular Biology Reports, 50(1). https://doi.org/10.1007/s11033-022-08015-7
Collins, R. A., & Cruickshank, R. H. (2012). The seven deadly sins of DNA barcoding. Molecular Ecology Resources, 13(6), n/a-n/a. https://doi.org/10.1111/1755-0998.12046
Yang, C., Zheng, Y., Tan, S., Meng, G., Rao, W., Yang, C., Bourne, D. G., O’Brien, P. A., Xu, J., Liao, S., Chen, A., Chen, X., Jia, X., Zhang, A., & Liu, S. (2020). Efficient COI barcoding using high throughput single-end 400 bp sequencing. BMC Genomics, 21(1). https://doi.org/10.1186/s12864-020-07255-
Zhang, H., & Bu, W. (2022). Exploring Large-Scale Patterns of Genetic Variation in the COI Gene among Insecta: Implications for DNA Barcoding and Threshold-Based Species Delimitation Studies. Insects, 13(5), 425. https://doi.org/10.3390/insects13050425